Biomedical Information Extraction
Uncovering Biomedical Data Silos Currently Locked Within Literature
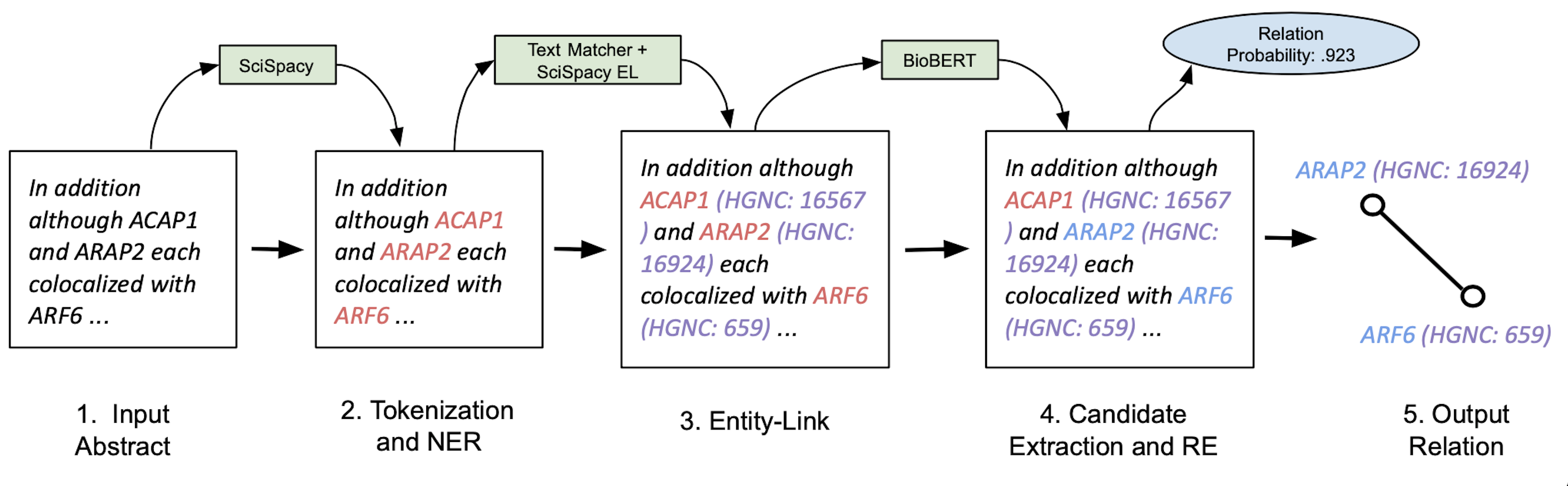 Overview:
Hundreds of thousands of papers are published each year in the biomedical domain which contain novel relationships between biomedical entities such as proteins and genes. However, with the number of articles that are being published it is impossible for any one person or structured database to comb through all this information to extract all the importnat relationships between entities mentioned in the text. Hence, it would be usesful to extract all of the relationship information contained in these articles as it contains vastly useful data.
Overview:
Hundreds of thousands of papers are published each year in the biomedical domain which contain novel relationships between biomedical entities such as proteins and genes. However, with the number of articles that are being published it is impossible for any one person or structured database to comb through all this information to extract all the importnat relationships between entities mentioned in the text. Hence, it would be usesful to extract all of the relationship information contained in these articles as it contains vastly useful data.
For instance, data concerning the relationship between biomedical entities can be used to augment a biomedical knowledge graph to add new nodes and edges that can increase the predictive power of graph learning algorithms that seek to identify new drug targets and disease treatments.
Hence during my time at OccamzRazor, I’ve been focusing on building a natural langauge processing(NLP) pipeline that is able to perform biomedical information extraction at a high level. As biomedical text is notorious for being very difficult to decode and understand, it is important that any information extraction pipeline in this domain be robust and generalizable. To this end, the pipeline I have engineered makes use of state of the art NLP models such as SciSpacy and BioBERT that we have augmented to be better suited for biomedical texts. Specifically, our pipeline consists of named entity recognition (NER), entity resolution (ER), and relation extraction(RE) where we have trained each individual model to perform at a high level and allow us to extract all the positive relationships betweeen biomedical entities contained in a given article. We then were able to run this novel pipeline over millions of articles in the PubMed corpus to extract thousands of protein and gene relationships that previously were locked away in these pieces of literature.
With this newly constructed dataset we have been able to augment a biomedical knowledge graph to provide lifts in better understand drug efficacy and providing novel disease treatments.